ORIGINAL ARTICLE
Practical and cost-effective method for the isolation of pollen grains from various sources
1
North Carolina State University, Department of Population Health and Pathobiology, College of Veterinary
Medicine, 1060 William Moore Drive, Raleigh, NC 27607, USA
2
United States Geological Survey, Florence Bascom Geoscience Center, 12201 Sunrise Valley Dr,
Reston VA 20192, USA
Submission date: 2024-07-30
Acceptance date: 2025-04-30
Online publication date: 2025-06-27
Acta Palaeobotanica 2025; 65(1): 122-129
HIGHLIGHTS
- Practical, cost-effective method for isolating pollen grains for mock standards
- Vacuum filtration effectively removes debris without lysing pollen grains
- Beneficial for palynology, forensic science, and environmental monitoring
- Method is tested using 25 species - herbarium, commercial, and fresh source materials
KEYWORDS
ABSTRACT
Mock standards, with known concentrations and varied characteristics, when analyzed alongside
unknown samples, can provide evaluation, optimization, and validation of scientific methods. Due to the scarcity
of commercially available pollen grains, this study introduces a practical and cost-effective method for isolating
pollen grains from various sources to be used in a mock pollen standard. Our method was tested using 25 diverse
species derived from different sources, including herbarium materials (n, 20; dated from 1941 to 2006), commercially
sourced (n, 2), and fresh hand-collected (n, 3), representing a wide range of taxonomic diversity and pollen
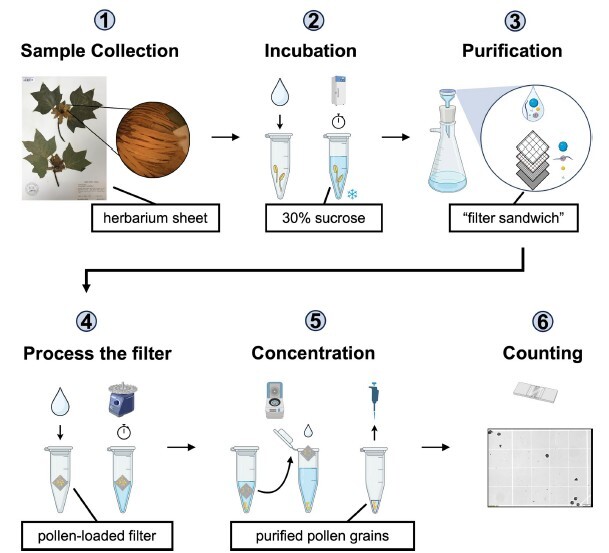
morphology. Isolation with vacuum filtration, which can be completed in a basic laboratory, easily removes inorganic
and organic debris while avoiding lysis of the pollen grains. This paper details the key steps in this method,
including a) collecting suitable plant materials containing pollen grains from fresh and herbarium specimens and
b) isolating, quantifying and storing the pollen grains. This approach is particularly beneficial for researchers in
palynology, plant biology, forensic science and environmental monitoring, offering a practical way to isolate pollen
grains for inclusion as a mock standard while preserving both morphological features and genetic material.
FUNDING
This research was sponsored by the
Army Research Office and was accomplished under
Cooperative Agreement Number W911NF-21-2-0129
REFERENCES (26)
1.
Ali, A.M.S., Rooney, P., Hawkins, J.A., 2022. Automatically counting pollen and measuring pollen production in some common grasses. Aerobiologia 38(4), 433–455. https://doi.org/10.1007/s10453....
2.
Banchi, E., Ametrano, C.G., Tordoni, E., Stanković, D., Ongaro, S., Tretiach, M., Pallavicini, A., Muggia, L., Verardo, P., Tassan, F., Trobiani, N., Moretti, O., Borney, M.F., Lazzarin, S., 2020. Environmental DNA assessment of airborne plant and fungal seasonal diversity. The Science of the Total Environment 738(140249), 140249. https://doi.org/10.1016/j.scit....
3.
Bell, K.L., Burgess, K.S., Botsch, J.C., Dobbs, E.K., Read, T.D., Brosi, B.J., 2019. Quantitative and qualitative assessment of pollen DNA metabarcoding using constructed species mixtures. Molecular Ecology 28(2), 431–455. https://doi.org/10.1111/mec.14....
4.
Bell, K.L., Burgess, K.S., Okamoto, K.C., Aranda, R., Brosi, B.J., 2016. Review and future prospects for DNA barcoding methods in forensic palynology. Forensic Science International. Genetics 21, 110–116. https://doi.org/10.1016/j.fsig....
5.
Devriese, A., Peeters, G., Brys, R., Jacquemyn, H., 2024. The impact of extraction method and pollen concentration on community composition for pollen metabarcoding. Applications in Plant Sciences 12(5), e11601. https://doi.org/10.1002/aps3.1....
6.
Duhoux, E., 1982. Mechanism of exine rupture in hydrated taxoid type of pollen. Grana 21(1), 1–7. https://doi.org/10.1080/001731....
7.
Erkens, R.H.J., Cross, H., Maas, J.W., Hoenselaar, K., Chatrou, L.W., 2008. Assessment of age and greenness of herbarium specimens as predictors for successful extraction and amplification of DNA. Blumea – Biodiversity, Evolution and Biogeography of Plants 53(2), 407–428. https://doi.org/10.3767/000651....
8.
Forster, M., Flenley, J.R., 1993. Pollen isolation and fractionation by equilibrium density gradient centrifugation. Palynology 17(1), 137–155. https://doi.org/10.1080/019161....
9.
Herbarium Sample Collection Protocol. Fit.edu. Available from: https://research.fit.edu/media.... Accessed 28 January 2024.
10.
Kakui, H., Tsurisaki, E., Sassa, H., Moriguchi, Y., 2020. An improved pollen number counting method using a cell counter and mesh columns. Plant Methods 16(1), 124. https://doi.org/10.1186/s13007....
11.
Kasai, Y., Leipe, C., Saito, M., Kitagawa, H., Lauterbach, S., Brauer, A., Tarasov, P.E., Goslar, T., Arai, F., Sakuma, S., 2021. Breakthrough in isolation of fossil pollen for dating of sediments by a new large-particle on-chip sorter. Science Advances 7(16), eabe7327. https://doi.org/10.1126/sciadv....
12.
Keller, A., Danner, N., Grimmer, G., Ankenbrand, M., von der Ohe, K., von der Ohe, W., Rost, S., Härtel, S., Steffan-Dewenter, I., 2015. Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biology (Stuttgart, Germany) 17(2), 558–566. https://doi.org/10.1111/plb.12....
13.
Kraaijeveld, K., de Weger, L.A., Ventayol García, M., Buermans, H., Frank, J., Hiemstra, P.S., den Dunnen, J.T., 2015. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Molecular Ecology Resources 15(1), 8–16. https://doi.org/10.1111/1755-0....
14.
Kelley, L., Rose, E., McCullough, B., Martinez, M., Baudelet, M., 2020. Non-destructive DNA analysis of single pollen grains. Forensic Chemistry 20, 100275. https://doi.org/10.1016/j.forc....
15.
Lang, D., Tang, M., Hu, J., Zhou, X., 2019. Genome-skimming provides accurate quantification for pollen mixtures. Molecular Ecology Resources 19(6), 1433–1446. https://doi.org/10.1111/1755-0....
17.
Pappas, C.S., Tarantilis, P.A., Harizanis, P.C., Polissiou, M.G., 2003. New method for pollen identification by FT-IR spectroscopy. Applied Spectroscopy 57(1), 23–27. https://doi.org/10.1366/000370....
18.
Parducci, L., Suyama, Y., Lascoux, M., Bennett, K.D., 2005. Ancient DNA from pollen: a genetic record of population history in Scots pine. Molecular Ecology 14(9), 2873–2882. https://doi.org/10.1111/j.1365....
19.
Rao, A.N., Ong, E.T., 1972. Germination of compound pollen grains. Grana 12(2), 113–120. https://doi.org/10.1080/001731....
20.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J.-Y., White, D.J., Hartenstein, V., Eliceiri, K., Tomancak, P., Cardona, A., 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9(7), 676–682. https://doi.org/10.1038/nmeth.....
21.
Shepherd, L.D., 2017. A non-destructive DNA sampling technique for herbarium specimens. PLoS One 12(8), e0183555. https://doi.org/10.1371/journa....
22.
Sikoparija, B., Galán, C., Smith, M., EAS QC Working Group, 2017. Pollen monitoring: between analyst proficiency testing. Aerobiologia 33(2), 191–199. https://doi.org/10.1007/s10453....
23.
Siriwattanakul, U., Piboonpocanun, S., Songnuan, W., 2019. Rapid pollen rupture and release of pollen cytoplasmic granules upon hydration of allergenic grass and weed species commonly found in subtropical regions. Aerobiologia 35(4), 719–730. https://doi.org/10.1007/s10453....
24.
Swenson, S.J., Gemeinholzer, B., 2021. Testing the effect of pollen exine rupture on metabarcoding with Illumina sequencing. PLOS ONE 16(2), e0245611. https://doi.org/10.1371/journa....
25.
Villagómez, G.N., Brachvogel, R.-C., Kárpáti, Z., Leonhardt, S.D., Schmitt, T., Ruedenauer, F.A., 2023. A common protocol for reliable comparison of pollen fatty acid profiles: highlighting pitfalls and proposing a methodology for ecological research. Frontiers in Ecology and Evolution 11, 1141832. https://doi.org/10.3389/fevo.2....
26.
Willard, D.A., Bernhardt, C.E., Weimer, L., Cooper, S.R., Gamez, D., Jensen, J., 2004. Atlas of pollen and spores of the Florida Everglades. Palynology 28(1), 175–227. https://doi.org/10.2113/28.1.1....
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.